An Accurate Mass Defect-based labeling strategy for Quantitative Proteomics
Zhong et al (2019) wrote about their development of an accurate mass-defect based labelling strategy for MS1-centric quantification in a recent Analytical Chemistry paper.
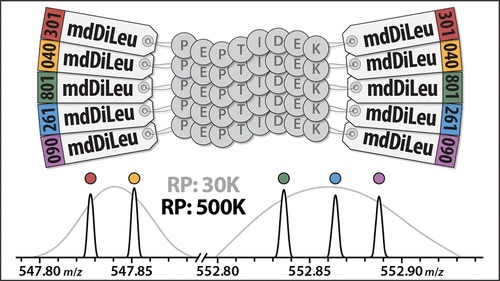
Specifically, researchers developed 5-plex mass defect N, N-dimethyl leucine (mdDiLeu) tags. These tags have multiple benefits; they can aid in the quantification of biological samples and have increased multiplexing due to the addition of mass difference isotopologues. Additionally, the synthesis of these cost effective tags is straightforward and only requires one reaction step, which can be done in any lab. Also, this mass defect-based labelling strategy is more accurate than isobaric label-based reporter ion quantification, as the latter is impacted by ratio compression.
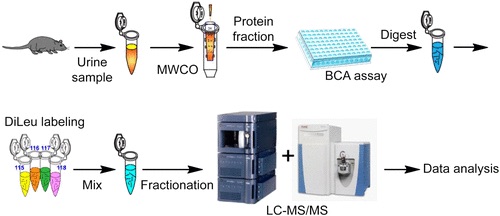
In this paper, Zhong et al (2019) demonstrate the efficacy of 5-plex mdDiLeu tags for quantitative proteomics by conducting mass spectrometry experiments with these tags on labelled Saccharomyces cerevisiae lysate digest.