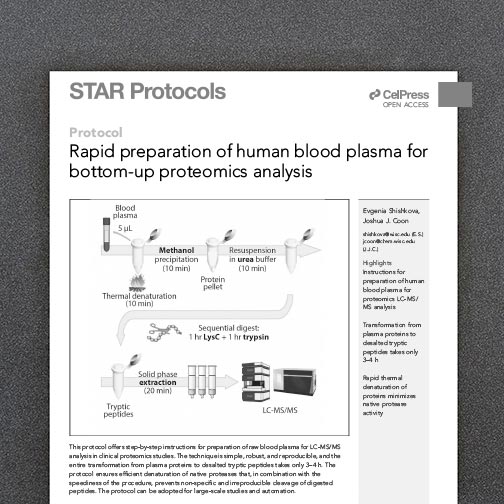
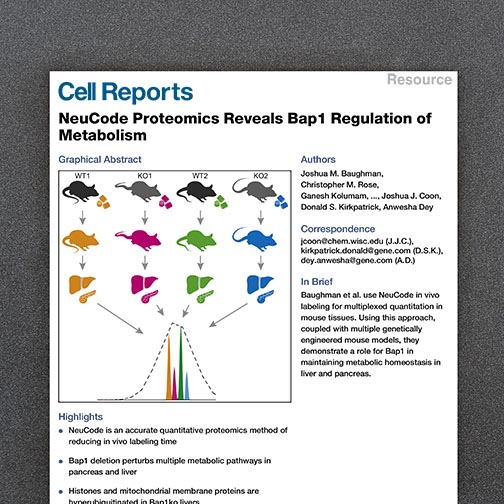
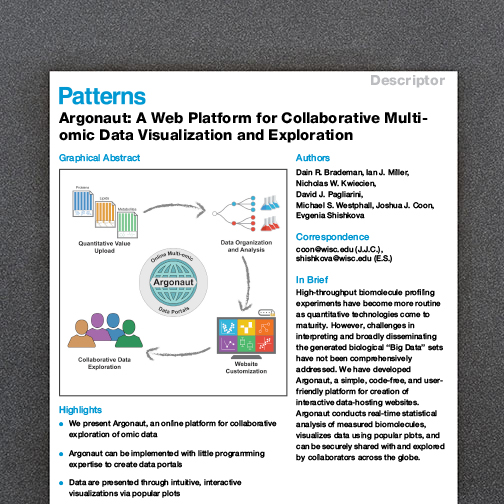
A major advantage of the -omics movement is the capacity to sample many features in a single experiment, establishing a global snapshot of the molecules that govern biology. Still, a fundamental challenge remains in establishing the biological relevance for features identified in these large-scale screens.
Additional resources can also be found at the Biomedical Technology Resources portal, an NIH innovative technology resource.
When appropriate, please acknowledge our support in publications and presentations: “This work was supported in part by the National Institutes of Health grant P41GM108538.”