Polo-like kinase 4 maintains integrity of centriolar satellites
RA Denu et al recently published a paper on the importance of polo-like kinase 4 for centriolar satellite integrity in the Journal of Biological Chemistry.
Polo-like kinase 4 (PLK4), a Ser/Hr protein kinase, is the “master regulator” of centriole duplication and can also play a role in centrosome function. Centrioles are cell organelles which are responsible for cell division. In addition, centrioles are housed in other organelles, called centrosomes.
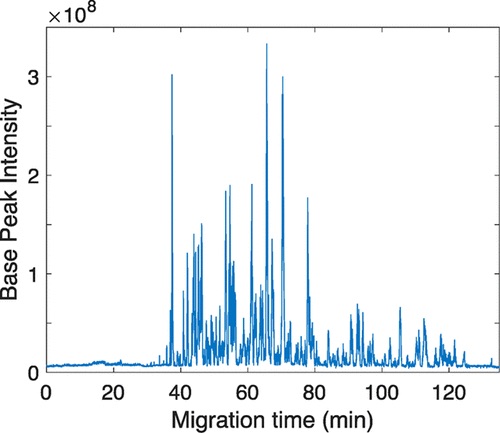
This study was an attempt to identify additional proteins regulated by PLK4. To do this, scientists generated an RPE-1 human cell line and genetically engineered analog sensitive PLK4As.
Scientists found that Ser-78 is important for maintaining the integrity of centriolar satellites, as this is where PLK4 phosphorylates CEP131. Ser-78 in centrosomal protein 131 is a direct substrate of PLK4.
Another finding is that inhibiting PLK4 or using a nonphosphorylatable CEP131 tended to result in “dispersed” centriolar satellites.