Li lab project uses DiLeu isobaric labeling to monitor neurotoxicity in chemotherapy in children
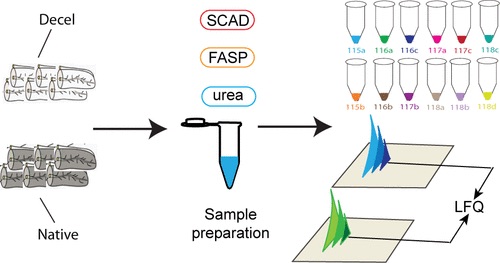
A new project by the Lingjun Li lab explores the use of the 4-plex N,N-dimethyl leucine (DiLeu) isobaric labeling strategy for protein identification and quantification in cerebrospinal fluid in children undergoing chemotherapy. The results from this and future studies will provide a means to monitor neurotoxicity and develop strategies to prevent central nervous system injury in response to chemotherapy in children.
The publication is Isobaric labeling strategy utilizing 4-plex N,N-dimethyl leucine (DiLeu) tags reveals proteomic changes induced by chemotherapy in cerebrospinal fluid of children with B-cell acute lymphoblastic leukemia from the Journal of Proteome Research.